Welcome to DNetPRO algorithm’s documentation!¶
Official implementation of the DNetPRO algorithm published on BioRXiv by Curti et al. The DNetPRO algorithm produces multivariate signatures starting from all the couples of variables analyzed by a Discriminant Analysis. The method is particularly designed to gene-expression data analysis and it was tested against the most common feature selection techniques. In the current implementation the DNetPRO object is totally equivalent to a scikit-learn feature-selection method and thus it provides the member functions fit (to train your model) and predict (to test a trained model on new samples). The combinatorial evaluation is performed using a C++ version of the code wrapped using Cython.
Overview¶
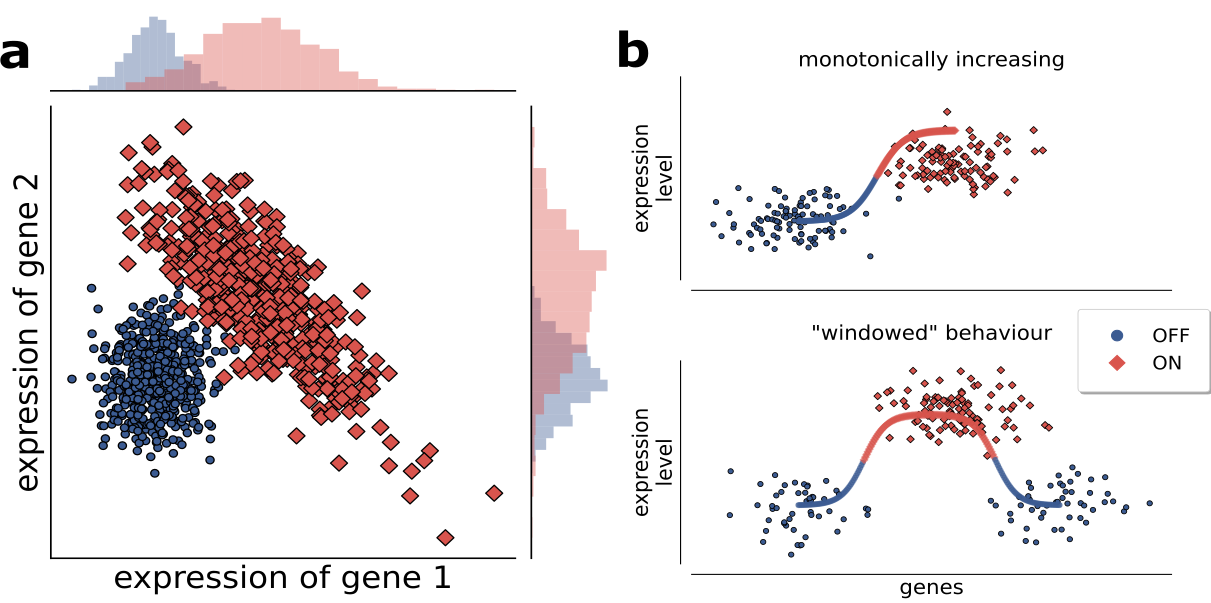
Methods that select variables for multi-dimensional signatures based on single-variable performance can have limits in predicting higher-dimensional signature performance. As shown in the Figure (a), in which both variables taken singularly perform poorly, but their performance becomes optimal in a 2-dimensional combination, in terms of linear separation of the two classes.
It is known that complex separation surfaces characterize classification tasks associated to image and speech recognition, for which Deep Networks are used successfully in recent times, but in many cases biological data, such as gene or protein expression, are more likely characterized by a up/down-regulation behavior (as shown in Figure (b) top), while more complex behaviors (e.g. a optimal range of activity, Figure (b) bottom) are much less likely. Thus, discriminant-based methods (and logistic regression methods alike) can very likely provide good classification performances in these cases (as demonstrated by our results with DNetPRO) if applied in at least two-dimensional spaces. Moreover, the of these methods (that generate very simple class separation surfaces, i.e. linear or quadratic) guarantee that a of a signature based on lower-dimensional signatures is feasible.
This consideration are relevant in particular for microarray data where we face on a small number of samples compared to a huge amount of variables (gene probes). This kind of problem, often called problem (where N is the number of features, i.e variables, and S is the number of samples), tend to be prone to overfitting and they are classified to ill-posed. The difficulty on the feature extraction can also increase due to noisy variables that can drastically affect the machine learning algorithms. Often is difficult to discriminate between noise and significant variables and even more as the number of variables rises.
In this project we propose a new method of features selection - DNetPRO, Discriminant Analysis with Network PROcessing - developed to outperform the mentioned above problems. The method is particularly designed to gene-expression data analysis and it was tested against the most common feature selection techniques. The method was already applied on gene-expression datasets but my work focused on the benchmark of it and on its optimization for Big Data applications. The pipeline algorithm is made by many different steps and only a part of it was designed to biological application: this allow me to apply (part of) the same techniques also in different kind of problems with good results (see Mizzi2018).
Usage example¶
You can use the DNetPRO algorithm into pure-Python modules or inside your C++ application.
C++ example¶
The easiest usage of DNetPRO algorithm is given by the example provided in the example_cpp folder. This script includes an easy-to-use command line to run the DNetPRO algorithm on a dataset stored into a file.
./bin/DNetPRO_couples
Usage: ./DNetPRO_couples -f <std :: string> -o <std :: string> [-frac <St16remove_referenceIfE> ] [-bin <St16remove_referenceIbE> ] [-verbose <St16remove_referenceIbE> ] [-probeID <St16remove_referenceIbE> ] [-nth <St16remove_referenceIiE> ]
DNetPRO couples evaluation 2.0
optional arguments:
-f, --input Input filename
-o, --output Output filename
-s, --frac Fraction of results to save
-b, --bin Enable Binary output
-q, --verbose Enable stream output
-p, --probeID ProbeID name to skip (true/false)
-n, --nth Number of threads to use
If you are interested in using DNetPRO algorithm inside your code you can simply import the dnetpro_couples.h and call the dnetpro_couples function.
Then all the results will be stored into a easy-to-manage score object.
Python example¶
The DNetPRO object is totally equivalent to a scikit-learn feature-selection method and thus it provides the member functions fit (to train your model) and predict (to test a trained model on new samples).
First of all you need to import the DNetPRO modules and then simply call the training/testing functions.
import pandas as pd
from DNetPRO import DNetPRO
from sklearn.naive_bayes import GaussianNB
from sklearn.model_selection import train_test_split
X = pd.read_csv('./example/data.txt', sep='\t', index_col=0, header=0)
y = np.asarray(X.columns.astype(float).astype(int))
X = X.transpose()
X_train, X_test, y_train, y_test = train_test_split(X.values, y, test_size=0.33, random_state=42)
dnet = DNetPRO(estimator=GaussianNB(), n_jobs=4, verbose=True)
Xnew = dnet.fit_transform(X_train, y_train)
print('Best Signature: {}'.format(dnet.get_signature()[0]))
print('Score: {:.3f}'.format(dnet.score(X_test, y_test)))